Continuing on one of the previous posts about data generation in python, next natural step in the analytic procedure is the Procrustes superimposition. Since this procedure enables direct analyses of configurations` shape, and all subsequent explorative visualizations, it must be employed first. This post concerns with the basic superimposition procedure, involving only two landmark configurations, i.e. two generated monsters. One monster will be used as a reference and one as a target, which should undergo superimposition. First steps in data genration are the same as before, with the exception of polarRotator function that can take any numpy array and reorder it according to polar rotation angle. This is very useful since generated landmarks are not ordered properly and do not “feel natural” in plots and analyses.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 | |
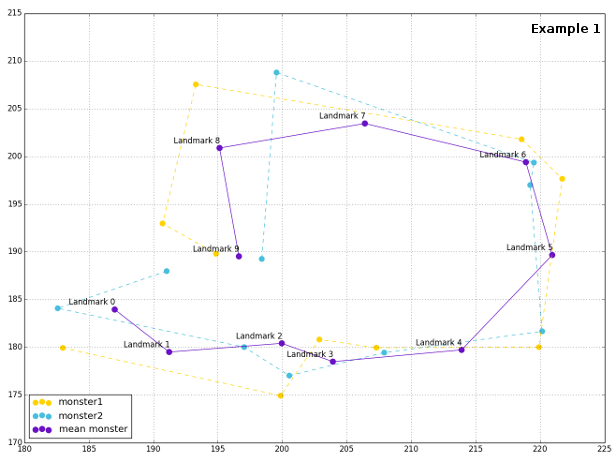
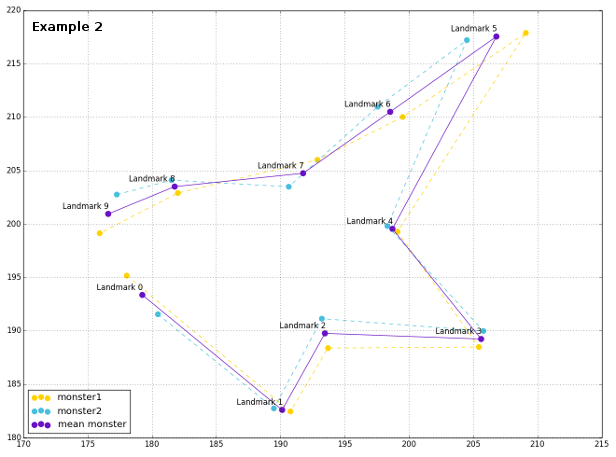
The plot of configurations reveals their spatial relationship, as well as the general mean shape. This time, since landmarks are ordered properly, one line would be enough for representing mean shapes, and shapes of respective configurations.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 | |
Procrustes superimposition revolves around three features of shape extraction, that is invariance of landmark configurations to position, scale and rotation. There are a number of excelent textbooks about the mathematics and logic, as well as procedures for Procrustes superimposition (Bookstein, 19911, Dryden and Mardia, 19982, Zelditch et al., 20123), but for this post direct inspiration was Morphometrics in R (Claude, 2008), especially with the basic procedure presented in the following function definition.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 | |
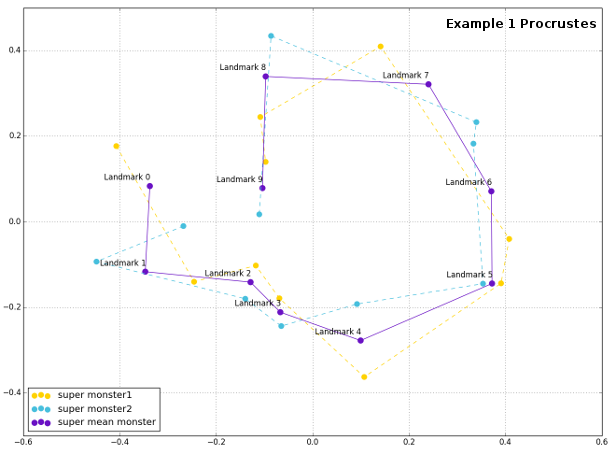
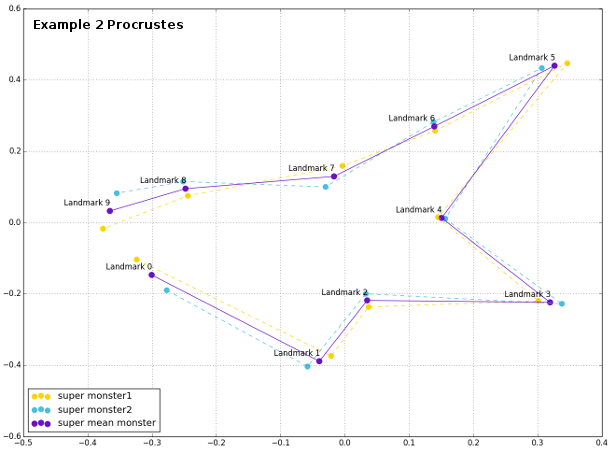
Plot of the superimposed configurations reveals that the Procrustes python was really able to force monsters` shape be more similar, removing the effects of orientation, size and rotation.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | |
Following posts should continue on this one and describe how the partial Procrustes superimposition for multilple configurations can be performed with fabulous sientific python.
-
Bookstein, F.L. 1991. Morphometric tools for landmark data: Geometry and Biology. Cambridge University press, Cambridge, UK.↩
-
Dryden, I.E. and K.V. Mardia. 1998. Statistical shape analysis. Wiley, Chichester, UK.↩
-
Zelditch, M.L., Swidersk, D.L. and H.D. Sheets. 2012. Geometric morphometrics for biologists: A primer. Second edition, Elsevier. ↩