Procrustes superimposition is the first analytic step in geometric morphometrics and this post shows one possible solution to performing it in Python1, using several functions defined in previous posts. First steps include data generation and definition of functions important for extracting shape variables from randomly generated landmark data. These functions are centsize which calculates centroid size from a numpy array (xy data) and transScale which translates landmark coordinates to the origin of the coordinate system and scales them to unit centroid size. The mshapr function is needed in the main superimposition function since it calculates succesive mean shapes (all configurations), excluding the configuration that is currently being rotated.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 | |
Definition of the pProc function follows the same rules as in the previous post, with the robust (and ugly for now) if clause that is only included to allow either pandas DataFrame or a numpy array as the input data (both for configurations and mean shape objects). It also returns only the rotated matrix, landmark configuration (pmat1) and not the mean shape.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 | |
Finally, the pGP function, that can perform partial Procrustes superimposition for any number of individuals (landmark configurations) and landmarks. Generated data has 200 individuals and 10 2D landmarks. For now this function will ask such information in the function call, so that mmat1 is pandas DataFrame with x, y, coordinates and individuals columns (generated above), numind is the number of individuals, dim is 2D (3D not yet supported), and numland is the number of landmarks. For clearer understanding of the following code, comments are included where appropriate.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 | |
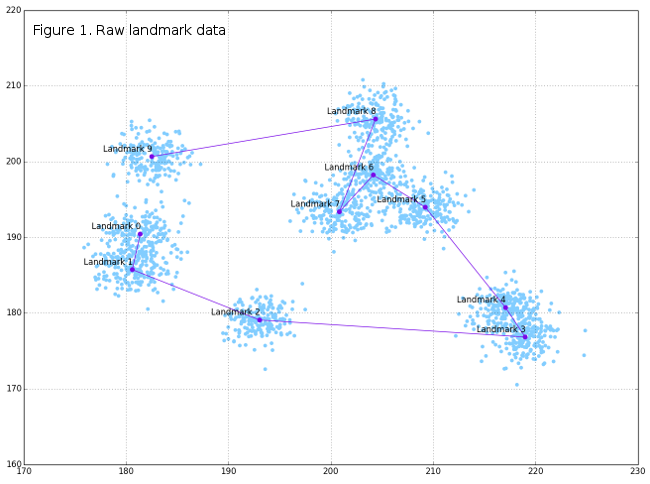
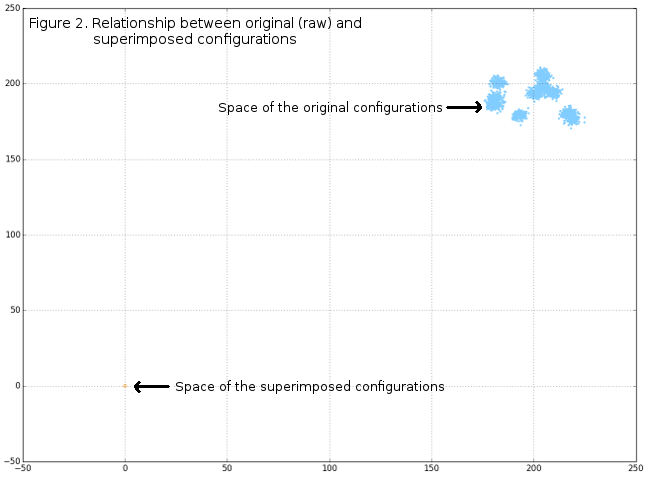
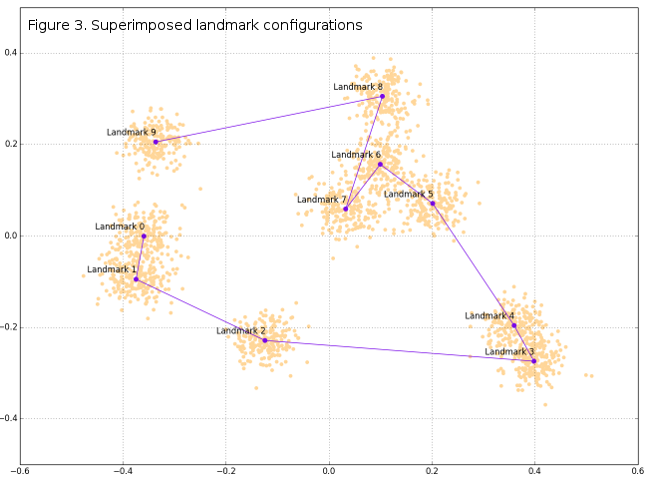
DataFrame tempRot holds the superimposed configurations (shape variables) that can, subsequently, be used in standard GM analyses and further. Of course, graphical display of superimposition results can be very interesting, and in this post only basic plots will be given, while some of the further posts may include more visualizations. Figure 1 shows the original raw-generated data, Figure 2 the spatial relationships between raw and superimposed data, while Figure 3 shows only superimposed data.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 | |
-
If using IPython (:)) best way is to paste code with the %paste magic function.↩